My current research interests include:
- Structural characterization of proteoglycans and protein:carbohydrate binding partners involved in immunology and infectious disease.
- Differentiation of Protein:Protein and Protein:Ligand complex conformations including proteomics as it applies to analysis of viral recruitment of the human ribosome/eukaryotic initiation factors and mapping of the protein modifications of the ribosomal protein subunits.
- Investigation and characterization of inflammatory protein:
glycosaminoglycan binding partners in rheumatoid arthritis.
Glycochemistry in immunology and infectious disease
Research currently being conducted in my group includes the use of ion trap mass spectrometry in combination with collision induced dissociation (CID) and Ion Mobility MS (IMS) for probing molecular structure and conformational changes. These techniques are used in the development of novel methodology for the characterization of carbohydrate structure in a variety of biological systems. My group and I have developed several software programs that allow for carbohydrate sequencing and compositional analysis in tandem with new methods that we developed for structurally characterizing proteoglycans. This research has advanced now to the area of protein-carbohydrate interactions in which we are analyzing chemokine-glycosaminoglycan non-covalent complexes involved in chemotaxis and inflammation.
Specific goals include sequencing and compositional analysis of heparin/heparan sulfate derived from the extracellular matrix of various healthy and diseased cells specifically as it relates to structure-function relationship. Particular emphasis is on GAG:Chemokine, Chemokine:G-protein coupled receptor and GAG:Chemokine:Receptor interactions.
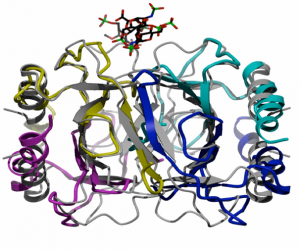
Chemokines, small chemotactic proteins, have been shown to bind GAGs both in vitro and in vivo, and recently it was demonstrated that this interaction is required for their function in vivo. Immobilization of chemokines on GAGs is thought to enhance their local concentration and facilitate the formation of chemokine gradients to guide the migration of cells, especially under flow conditions. GAG-binding may also help recruit chemokines to specific populations of cells, thereby contributing to the control of cell migration in a receptor independent way. An emerging hypothesis is that glycosaminoglycans (GAGs) play a role in the in vivo function of chemokines, and that specificity in these interactions could alter the apparent redundancy of the chemokine:receptor interaction. In this particular area of research we are developing and utilizing mass spectrometric techniques that will allow us to analyze the non-covalent complexes of GAG-Chemokine, Chemokine-Receptor and eventually GAG-Chemokine-Receptor assemblies. (At left is a crystal structure we solved showing the binding site of the GAG drug, Arixtra bound at the dimer interface of eotaxin, a known chemokine upregulated in asthma.) Specific questions we will address include:
- Do different chemokines bind GAG’s with different affinity?
- What role does multimerization play in GAG binding?
- Is there GAG sequence specificity in binding?
- What are the structures of the GAG:Chemokine complexes?
- How do these interactions affect inflammation and disease in vivo?
- What, if any, conformational differences of the GAG or Chemokine are critical to binding?
Ribosome recruitment by viruses and attending protein modifications
The ribosome, the cellular catalyst of protein synthesis, is one of the largest and most complex biological macromolecules. We are currently involved in a collaborative research project to test the hypothesis that the ribosome and its component subunits exist in multiple functional states in human cells. Preliminary evidence suggests that distinct forms of the ribosome, differing by one or a few component proteins, may play critical roles in the control of gene expression during viral infection. Upon infection by viruses such as hepatitis C, an internal ribosome entry site (IRES) RNA binds to host cell ribosomes and recruits them for viral protein synthesis. Ribosome-IRES complexes are separated from ribosomes incapable of IRES recognition, and the components of each sample analyzed by ESI-FTICR and HD-MS Ion Mobility mass spectrometry.
These MS methods are used to probe protein complexes that comprise mammalian ribosomes, and the accompanying initiation factors, both before and after infection with Hepatitis C virus. The post-translational modifications found to protein complexes have been extremely interesting, particularly those showing phosphorylation and sulfation. One of our recent manuscripts published in Nature Methods, describes a first time protocol for detecting tyrosine sulfation and determining the temporal resolution of sulfation on N-terminal GPCR. As part of this project, I installed a new design mass spectrometer (Waters Synapt HDMS), capable of detecting megadalton protein complexes. This particular instrument was the first of its kind installed in the United States. This instrument has allowed for the analysis of protein:protein and protein:carbohydrate binding partners. The eukaryotic initiation factor 3, shown above, is one such complex with a mass of 800,000 Da which we have successfully analyzed and determined the subunit interactions.
Experiments are underway which involve denaturing the human ribosome complexes and analyzing the constituent proteins by ion trap and IM mass spectrometry using both a bottom up and top down approach. Data thus far collected indicate all 32 proteins from both the native and IRES recruited 40 S ribosome have been identified; many contain various post-translational modifications. The eIF3 complex of the IRES recruited ribosome has also been investigated and sites of phosphorylation have been unambiguously identified. Equally important is the fact that we have probed the interaction of the various subunits and are now looking at conformational differences/folding and unfolding during complex disruption. Technology developed in the course of this project will be valuable to the study of many macromolecular complexes that play central roles in the control of gene expression.